|
林勇欣 副教授 國立陽明交通大學 生物資訊及系統生物研究所 Yeong-Shin Lin, Associate Professor Institute of Bioinformatics and Systems Biology National Yang Ming Chiao Tung University Hsinchu, Taiwan |
 |
| Main Page | Lab Members | Lectures | Resources | Bioinformatics Center |
Research InterestsOur major interest is in the processes and mechanisms of Evolution. We also use evolutionary concepts to study Genomics, Structural Biology, Systems Biology, Phylogenetics and Biogeography.
We use a wide range of research techniques, from the molecular to the organismal level, from DNA sequences to protein structures, from genomics to proteomics, and from experimental to computational studies, including developing Bioinformatic methods and tools, to study these problems. 客家電視《四問四吵》EP05:客家語言分布與基因遺傳 Publications
[e-journal] [PubMed] [IF] [Cited by] Pei-Ying Kao, Ming-Hui Chen, Wei-An Chang, Mei-Lin Pan, Wei-Der Shu, Yuh-Jyh Jong, Hsien-Da Huang, Cheng-Yan Wang, Hong-Yan Chu, Cheng-Tsung Pan, Yih-Lan Liu*, and Yeong-Shin Lin*. (2023). A genome-wide association study (GWAS) of the personality constructs in CPAI-2 in Taiwanese Hakka populations. PLoS ONE. 18: e0281903. [e-journal] [PubMed] [IF] [Cited by] 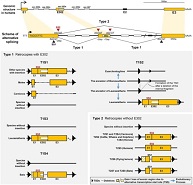 Cheng-Tsung Pan and Yeong-Shin Lin*. (2020).
MicroRNA retrocopies generated via L1-mediated retrotransposition in placental mammals help to reveal how their parental genes were transcribed.
Scientific Reports. 10: 20612.
Cheng-Tsung Pan and Yeong-Shin Lin*. (2020).
MicroRNA retrocopies generated via L1-mediated retrotransposition in placental mammals help to reveal how their parental genes were transcribed.
Scientific Reports. 10: 20612.[e-journal] [PubMed] [IF] [Cited by] Ho-Chang Kuo, Cheng-Tsung Pan, Ying-Hsien Huang, Fu-Chen Huang, Yeong-Shin Lin, Sung-Chou Li, and Lien-Hung Huang. (2019). Global Investigation of Immune Repertoire Suggests Kawasaki Disease Has Infectious Cause. Circulation Journal. 83: 2070-2078. [e-journal] [PubMed] [IF] [Cited by] Lien-Hung Huang, Ho-Chang Kuo, Cheng-Tsung Pan, Yeong-Shin Lin, Ying-Hsien Huang, and Sung-Chou Li*. (2018). Multiomics analyses identified epigenetic modulation of the S100A gene family in Kawasaki disease and their significant involvement in neutrophil transendothelial migration. Clinical Epigenetics. 10: 135. [e-journal] [PubMed] [IF] [Cited by] Yi-Hsueh Lee, Chia-Pei Chang, Yu-Ju Cheng, Yi-Yi Kuo, Yeong-Shin Lin, and Chien-Chia Wang*. (2017). Evolutionary gain of highly divergent tRNA specificities by two isoforms of human histidyl-tRNA synthetase. Cellular and Molecular Life Sciences. 74: 2663-2677. [e-journal] [PubMed] [IF] [Cited by] Min-Kung Hsu, I-Ching Wu, Ching-Chia Cheng, Jen-Liang Su, Chang-Huain Hsieh, Yeong-Shin Lin, and Feng-Chi Chen*. (2015). Triple-layer dissection of the lung adenocarcinoma transcriptome: regulation at the gene, transcript, and exon levels. Oncotarget. 6: 28755-28773. [e-journal] [PubMed] [IF] [Cited by] Chia-Pei Chang, Chih-Yao Chang, Yi-Hsueh Lee, Yeong-Shin Lin, and Chien-Chia Wang*. (2015). Divergent alanyl-tRNA synthetase genes of Vanderwaltozyma polyspora descended from a common ancestor through whole-genome duplication followed by asymmetric evolution. Molecular and Cellular Biology. 35: 2242-2253. [e-journal] [PubMed] [IF] [Cited by] Han Liang, Yeong-Shin Lin, and Wen-Hsiung Li*. (2008). Fast evolution of core promoters in primate genomes. Molecular Biology and Evolution. 25: 1239-1244. [e-journal] [PubMed] [IF] [Cited by] Yeong-Shin Lin, Wei-Lun Hsu, Jenn-Kang Hwang*, and Wen-Hsiung Li*. (2007). Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Molecular Biology and Evolution. 24: 1005-1011. [e-journal] [PubMed] [IF] [Cited by] 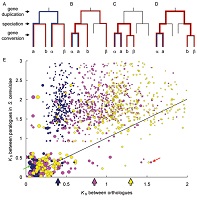 Yeong-Shin Lin, Jenn-Kang Hwang, and Wen-Hsiung Li*. (2007).
Protein complexity, gene duplicability and gene dispensability in the yeast genome.
Gene. 387: 109-117.
Yeong-Shin Lin, Jenn-Kang Hwang, and Wen-Hsiung Li*. (2007).
Protein complexity, gene duplicability and gene dispensability in the yeast genome.
Gene. 387: 109-117.[e-journal] [PubMed] [IF] [Cited by] Yeong-Shin Lin, Jake K. Byrnes, Jenn-Kang Hwang, and Wen-Hsiung Li*. (2006). Codon usage bias versus gene conversion in the evolution of yeast duplicate genes. Proc. Natl. Acad. Sci. USA. 103: 14412-14416. [e-journal] [PubMed] [IF] [Cited by]
[e-journal] [PubMed] [IF] [Cited by] 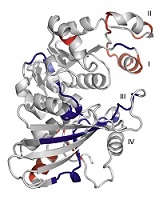 Yeong-Shin Lin*. (2008).
Using a strategy based on the concept of convergent evolution to identify residue substitutions responsible for thermal adaptation.
Proteins - structure, function, and bioinformatics. 73: 53-62.
Yeong-Shin Lin*. (2008).
Using a strategy based on the concept of convergent evolution to identify residue substitutions responsible for thermal adaptation.
Proteins - structure, function, and bioinformatics. 73: 53-62.[e-journal] [PubMed] [IF] [Cited by] [Data] Chih-Hao Lu, Yeong-Shin Lin, Yu-Ching Chen, Chin-Sheng Yu, Shi-Yu Chang, and Jenn-Kang Hwang*. (2006). The fragment transformation method to detect the protein structural motifs. Proteins - structure, function, and bioinformatics. 63: 636-643. [e-journal] [PubMed] [IF] [Cited by] Yu-Ching Chen, Yeong-Shin Lin, Chih-Jen Lin, and Jenn-Kang Hwang*. (2004). Prediction of the bonding states of cysteines using the Support Vector Machines based on multiple feature vectors and Cysteine State Sequences. Proteins - structure, function, and bioinformatics. 55: 1036-1042. [e-journal] [PubMed] [IF] [Cited by] 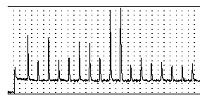 Yeong-Shin Lin*, Fu-Guo Robert Liu, Tzi-Yuan Wang, Cheng-Tsung Pan, Wei-Ting Chang, and Wen-Hsiung Li. (2011).
A simple method using PyrosequencingTM to identify de novo SNPs in pooled DNA samples.
Nucleic Acids Research. 39: e28.
Yeong-Shin Lin*, Fu-Guo Robert Liu, Tzi-Yuan Wang, Cheng-Tsung Pan, Wei-Ting Chang, and Wen-Hsiung Li. (2011).
A simple method using PyrosequencingTM to identify de novo SNPs in pooled DNA samples.
Nucleic Acids Research. 39: e28.[e-journal] [PubMed] [IF] [Cited by] [Pyrogram SNP Miner]  Chia-Hao Chang, Yi Ta Shao, Wen-Chung Fu, Kazuhiko Anraku, Yeong-Shin Lin, and Hong Young Yan*. (2015).
Differentiation of visual spectra and nuptial colorations of two Paratanakia himantegus subspecies (Cyprinoidea: Acheilognathidae) in response to the distinct photic conditions of their habitats.
Zoological Studies. 54: 43.
Chia-Hao Chang, Yi Ta Shao, Wen-Chung Fu, Kazuhiko Anraku, Yeong-Shin Lin, and Hong Young Yan*. (2015).
Differentiation of visual spectra and nuptial colorations of two Paratanakia himantegus subspecies (Cyprinoidea: Acheilognathidae) in response to the distinct photic conditions of their habitats.
Zoological Studies. 54: 43.[e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Fan Li, Kwang-Tsao Shao, Yeong-Shin Lin, Takahiro Morosawa, Sungmin Kim, Hyeyoung Koo, Won Kim, Jae-Seong Lee, Shunping He, Carl Smith, Martin Reichard, Masaki Miya, Tetsuya Sado, Kazuhiko Uehara, Sbastien Lavoué, Wei-Jen Chen*, and Richard L. Mayden. (2014). Phylogenetic relationships of Acheilognathidae (Cypriniformes: Cyprinoidea) as revealed from evidence of both nuclear and mitochondrial gene sequence variation: Evidence for necessary taxonomic revision in the family and the identification of cryptic species. Molecular Phylogenetics and Evolution. 81: 182-194. [e-journal] [PubMed] [IF] [Cited by] Hui-Yu Teng, Yeong-Shin Lin, and Chyng-Shyan Tzeng*. (2009). A new Anguilla species and a reanalysis of the phylogeny of freshwater eels. Zoological Studies. 48: 808-822. [e-journal] [PubMed] [IF] [Cited by]  Chyng-Shyan Tzeng*, Yeong-Shin Lin, Si-Min Lin, Tzi-Yuan Wang, and Feng-Yung Wang. (2006).
The phylogeography and population demographics of selected freshwater fishes in Taiwan.
Zoological Studies. 45: 285-297.
Chyng-Shyan Tzeng*, Yeong-Shin Lin, Si-Min Lin, Tzi-Yuan Wang, and Feng-Yung Wang. (2006).
The phylogeography and population demographics of selected freshwater fishes in Taiwan.
Zoological Studies. 45: 285-297.[e-journal] [PubMed] [IF] [Cited by] Yeong-Shin Lin*, Chyng-Shyan Tzeng, and Jenn-Kang Hwang*. (2005). Reassessment of morphological characteristics in freshwater eels (genus Anguilla, Anguillidae) shows congruence with molecular phylogeny estimates. Zoologica Scripta. 34: 225-234. [e-journal] [PubMed] [IF] [Cited by] Yeong-Shin Lin, Yu-Ping Poh, Si-Min Lin, and Chyng-Shyan Tzeng*. (2002). Molecular techniques to identify freshwater eels: RFLP analyses of PCR-amplified DNA fragments and allele-specific PCR from mitochondrial DNA. Zoological Studies. 41: 421-430. [e-journal] [PubMed] [IF] [Cited by] 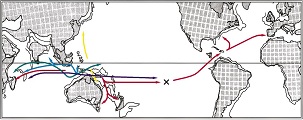 Yeong-Shin Lin, Yu-Ping Poh, and Chyng-Shyan Tzeng*. (2001).
Which is the dispersal route of the ancient Atlantic eels: Reanalysis of the phylogeny of freshwater eels.
Journal of Taiwan Fisheries Research. 9: 161-173.
Yeong-Shin Lin, Yu-Ping Poh, and Chyng-Shyan Tzeng*. (2001).
Which is the dispersal route of the ancient Atlantic eels: Reanalysis of the phylogeny of freshwater eels.
Journal of Taiwan Fisheries Research. 9: 161-173.[e-journal] [PubMed] [IF] [Cited by] Yeong-Shin Lin, Yu-Ping Poh, and Chyng-Shyan Tzeng*. (2001). A phylogeny of freshwater eels inferred from mitochondrial genes. Molecular Phylogenetics and Evolution. 20: 252-261. [e-journal] [PubMed] [IF] [Cited by] [Cited by The Eel ]
[e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Wei-Chuan Chiang, Yeong-Shin Lin, Nian-Hong Jang-Liaw, and Kwang-Tsao Shao*. (2016). Complete mitochondrial genome of the longfin mako shark, Isuru spaucus (Chondrichthyes, Lamnidae). Mitochondrial DNA. 27: 690-691. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Nian-Hong Jang-Liaw, Yeong-Shin Lin, Aaron Carlisle, Hua Hsun Hsu, Yun-Chih Liao, and Kwang-Tsao Shao*. (2016). The complete mitochondrial genome of the salmon shark, Lamna ditropis (Chondrichthyes, Lamnidae). Mitochondrial DNA. 27: 316-317. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Rima W. Jabado, Yeong-Shin Lin, and Kwang-Tsao Shao*. (2015). The complete mitochondrial genome of the sand tiger shark, Carcharias taurus (Chondrichthyes, Odontaspididae). Mitochondrial DNA. 26: 728-729. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Kwang-Tsao Shao, Yeong-Shin Lin, An-Yi Tsai, Pin-Xuan Su, and Hsuan-Ching Ho*. (2015). The complete mitochondrial genome of the shortfin mako, Isurus oxyrinchus (Chondrichthyes, Lamnidae). Mitochondrial DNA. 26: 475-476. [e-journal] [PubMed] [IF] [Cited by] Fan Li, Kwang-Tsao Shao, Yeong-Shin Lin, and Chia-Hao Chang*. (2015). The complete mitochondrial genome of the Rhodeus shitaiensis (Teleostei, Cypriniformes, Acheilognathidae). Mitochondrial DNA. 26: 301-302. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Kwang-Tsao Shao, Yeong-Shin Lin, Yi-Chiao Fang, and Hsuan-Ching Ho*. (2014). The complete mitochondrial genome of the great white shark, Carcharodon carcharias (Chondrichthyes, Lamnidae). Mitochondrial DNA. 25: 357-358. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Kwang-Tsao Shao, Yeong-Shin Lin, Hsuan-Ching Ho, and Yun-Chih Liao*. (2014). The complete mitochondrial genome of the big-eye thresher shark, Alopias superciliosus (Chondrichthyes, Alopiidae). Mitochondrial DNA. 25: 290-292. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Kwang-Tsao Shao, Yeong-Shin Lin, Wei-Chuan Chiang, and Nian-Hong Jang-Liaw*. (2014). Complete mitochondrial genome of the megamouth shark Megachasma pelagios (Chondrichthyes, Megachasmidae). Mitochondrial DNA. 25: 185-187. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Kwang-Tsao Shao, Yeong-Shin Lin, Yun-Chih Liao*. (2013). The complete mitochondrial genome of the three-spot seahorse, Hippocampus trimaculatus (Teleostei, Syngnathidae). Mitochondrial DNA. 24: 665-667. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Han-Yang Lin, Nian-Hong Jang-Liaw, Kwang-Tsao Shao, Yeong-Shin Lin, Hsuan-Ching Ho*. (2013). The complete mitochondrial genome of the tiger tail seahorse, Hippocampus comes (Teleostei, Syngnathidae). Mitochondrial DNA. 24: 199-201. [e-journal] [PubMed] [IF] [Cited by] Chia-Hao Chang, Nian-Hong Jang-Liaw, Yeong-Shin Lin, Yi-Chiao Fang, and Kwang-Tsao Shao*. (2013). Authenticating the use of dried seahorses in the traditional Chinese medicine market in Taiwan using molecular forensics. Journal of Food and Drug Analysis. 21: 310-316. [e-journal] [PubMed] [IF] [Cited by] |
Contact
Lectures普通生物學(一)General Biology (I) 計算生物實驗 Computational Biology Lab. 遺傳學 Genetics 生物多樣性與生態 Biodiversity and Ecology 演化生物學 Evolutionary Biology 分子演化 Molecular Evolution LinksNCBIEnsEMBL Genome OnLine Database Approved Sequencing Targets UCSC Genome Bioinformatics Stanford Genomic Resources TGI - The Gene Index J. Craig Venter Institute Broad Institute HapMap SGD SMD MIPS RCSB PDB SCOP ExPASy - SwissProt - PROSITE CE - Combinatorial Extension RepeatMasker MEGA PAUP PAML PhyML CONSEL MacClade MrBayes DAMBE LiKaKs Structure (population) DnaSP Arlequin MCL - a cluster algorithm for graphs The R Manuals SimpleR Chi-square Test Fisher's Exact Test Kolmogorov-Smirnov Test Nature Science PNAS PLoS Biology Current Biology Cell EMBO Nature Ecology & Evolution Nature Genetics Nature Biotechnology Trends in Genetics Genome Research Genome Biology Molecular Biology & Evolution Nucleic Acids Research Genetics Evolution Bioinformatics Journal of Molecular Biology Journal of Molecular Evolution MPE Proteins Gene 國科會 Journal Citation Reports 政府電子採購網 |
| Main Page | Lab Members | Lectures | Resources | Bioinformatics Center |
 nycu.edu.tw
nycu.edu.tw